Quantitative PCR (or qPCR), also known as real-time PCR, was developed as a precise, efficient and rapid method for nucleic acid detection. It is a method in many laboratories for diagnostic and food applications. This technique is based on traditional Polymerase Chain Reaction (PCR) technology. It is a refinement of the original PCR created by Kary Mullis and with some improvements: (1) Quantitative PCR combines nucleic acid amplification and detection in one step. (2) It can use smaller amounts of starting material than traditional PCR. (3) It is possible to quantify the product based on fluorescent detection. (4) There is no post-amplification process. The instrument of qPCR is a thermal cycler. qPCR uses the linearity of DNA amplification to determine absolute or relative quantities of a known sequence in a sample. By using a fluorescent reporter in the reaction, DNA generation can be measured in the qPCR assay.
qPCR chemistries can be classified into two main groups: double-stranded DNA intercalating molecules (SYBR Green I or ethidium bromide), and fluorophore-labeled oligonucleotides. The former is the simplest and most cost-effective method as amplicon-specific labeled hybridization probes are not required. However, it is not specific since the dye binds to all double-stranded DNA formed during the PCR reaction. The latter can be further divided into three subgroups according to the type of fluorescent molecules used in the PCR reaction: (1) Primer-probes. (2) Probes, hydrolysis (TaqMan) and hybridization. (3) Analogues of nucleic acids.
SYBR Green 1 is a nonsequence-specific fluorogenic minor groove DNA-binding dye that intercalates into dsDNA but can not bind to single strand DNA. It could emit a strong fluorescent signal once bind to dsDNA (Fig. 1). The fluorescence signal increases during polymerization and decreases when DNA is denatured. Fluorescent measurements are performed at the end of the elongation step of each PCR cycle to determine the increasing amount of amplified DNA.
 Fig. 1 Primer and probe annealing and polymerization. (Arya M, et al. 2005)
Fig. 1 Primer and probe annealing and polymerization. (Arya M, et al. 2005)
A fluorogenic nonextendable ‘TaqMan’ probe and a forward and reverse primer are used in the real-time quantitative TaqMan assay. The efficiency of the assay is mainly dependent on 5' to 3' nuclease activity. So any enzyme with 5' nuclease activity can be applied here, mostly Taq polymerase. The oligonucleotide probe has a covalently bonded fluorescent reporter dye and quencher dye at the 5' and 3' ends respectively. Fluorescent reporter dyes like 6-carboxyfluorescein (FAM), tetrachloro-6-carboxyfluorescein (TET) and hexacholoro-6-carboxyfluorescein (HEX) are in use. Quenchers include 6-carboxytetramethylrhodamine (TAMRA) and 4-(dimethylaminoazo) benzene-4-carboxylic acid (DABCYL).
 Fig. 2 TaqMan assay. When the TaqMan probe exists, the reporter and quencher stay close to each other, and the fluorescence can't emit. After denaturation, the primer and TaqMan probe anneal to the complementary DNA strand. The 5' endonuclease activity of the Taq DNA polymerase cleaves the probe during the extension phase following hybridization. Then the reporter and quencher dyes are separated and fluorescence can be detected. (Arya M, et al. 2005)
Fig. 2 TaqMan assay. When the TaqMan probe exists, the reporter and quencher stay close to each other, and the fluorescence can't emit. After denaturation, the primer and TaqMan probe anneal to the complementary DNA strand. The 5' endonuclease activity of the Taq DNA polymerase cleaves the probe during the extension phase following hybridization. Then the reporter and quencher dyes are separated and fluorescence can be detected. (Arya M, et al. 2005)
Molecular beacons also contain covalently bound fluorescent and quenching dyes on the single-stranded DNA molecule. They are designed to fit the hairpin or stem-loop structure, while freeing in solution so that the fluorescent dye and the quencher are closely adjacent, so that fluorescence resonance energy transfer (FRET) occurs (Fig. 3). The hairpin structure consists of a stem and two loops, which are complementary to the target sequence. This configuration guarantees that the reporter fluorescent dye and the quencher are kept very close, so no fluorescence is detected. During the annealing process, the beacon hybridizes with the target sequence, thereby changing its conformation, isolating the reporter gene and the quenching dye, then fluorescence is emitted.
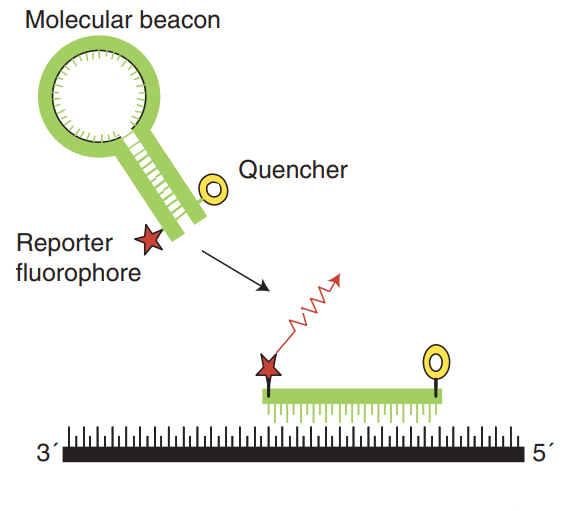 Fig. 3 Molecular beacons. (Arya M, et al. 2005)
Fig. 3 Molecular beacons. (Arya M, et al. 2005)
The qPCR results are recorded through interconnected computer graphics generated in the thermal cycler. Generally, four kinds of analysis are carried out: amplification curve, dissociation, curve, spectrum, and component. Through the amplification curve, the Ct value can be checked, which is crucial for the calculation of specificity and determination of the positive samples. If the value of Ct is above 35, it indicates false-positive results. The dissociation curve could reflect the emission of fluorescence by melting temperature (Tm). This analysis can verify non-specific amplification of primer dimers. Spectrum assessment shows the capture of fluorescence by certain filters. The component shows the fluorescence emission of each dye or reporter used in the reaction. Therefore, the background should keep constant and the employed dye should outrange it according to how many PCR cycles are performed. On the other hand, if a TaqMan probe is used, three parameters are important: the background, reporter, and light inhibitor signal (the quencher). The four parameters must be considered together. Optimal behavior of standard samples during the chemical reactions ensures the results of the other samples.
 Fig. 4 Model of a single amplification plot. (Arya M, et al. 2005)
Fig. 4 Model of a single amplification plot. (Arya M, et al. 2005)
References: